If you have any questions or suggestions, please contact us at yichengliu@zju.edu.cn.
Citation
If you use MAGPIE in your research, please cite the following paper:
Liu, Y., Zhang, T., You, N. et al. MAGPIE: accurate pathogenic prediction for multiple variant types using machine learning approach. Genome Med 16, 3 (2024). https://doi.org/10.1186/s13073-023-01274-4 BibTex EndNote RefManChange Log
v0.1.0
Update MAGPIE user interface.
Add up to 14 other prediction tools results.
Add up to 14 other prediction tools results.
v0.0.3
Update MAGPIE task query page.
v0.0.2
Add other prediction tools results. Add prediction results for variants in GRCh37/hg19 assembly.
v0.0.1
Add pathogenicity classification. Add ClinVar VUS prediction results.
Todo List
Add MAGPIE score for all possible exonic variants as soon as possible. Prediction is currently in progress.
Introduction
The model was trained to predict pathogenic scores of multi-type variants and included three steps. First, candidate variants were annotated with high-dimensional features covering 6 different modalities. Second, automatic feature engineering and separated feature selection were undertaken step by step. Finally, a gradient boosting method with controllable tuning was implemented to train the model and obtain predictions for the pathogenicity of variants.
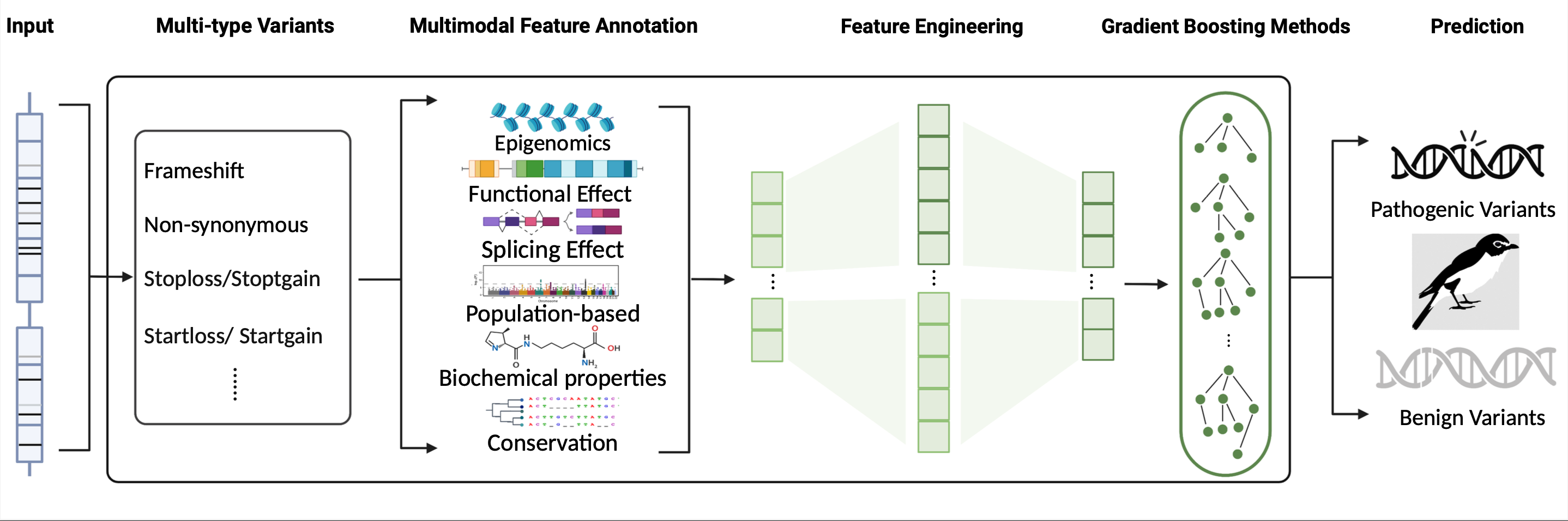
Preview
Preview
